Wasabi plugin system allows to add custom command-line tools to the Wasabi interface via plugin JSON API. Both the plugin system and included plugin files have been updated. See http://wasabiapp.org/plugins for more details. JSON API additions (“version”, “debug”, “disable”, “dirpath” option,…
Author: Andres
Wasabi updates 10.2018
Thanks for testing Wasabi and reporting issues on the Wasabi GitHub page. We’ve got progress: Added tree scalebar and domain restriction option for import URLs Fixed data export bugs together with numerous other reported issues (undo, rendering etc.) The latest Wasabi is…
Wasabi updates 8.2018
Wasabi has improvements in tree annotation and plugin support: Support for tree labels (e.g. bootstrap values) Faster tree import and rendering New URL parameters for customizing Wasabi (see http://wasabiapp.org/rest) Browse shared analysis library folders without Wasabi user account Updated Wasabi…
Wasabi updates 12.2017
The issue reporting for Wasabi has moved to the Wasabi GitHub page. Some of the improvements and bug fixes in recent Wasabi updates: Fixed issues with importing certain fasta headers. Fixed issue with importing tree and sequence from separate web…
Wasabi outage
Wasabi was offline 20.-21. March due to unexpected changes in wasabiapp.org nameserver settings. The issue has been resolved and hopefully stays that way.
Wasabi update 9.2016
We have released Wasabi maintenance update. Changes include: Wasabi can now import Ensembl EPO alignments crossing block boundaries Bug fixes and optimizations (e.g. improved Ensembl genetree support, server can provide data in jsonp format etc.) The updated Wasabi is ready…
Wasabi integrated to Ensembl genome browser
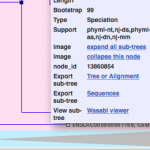
The world’s largest genome browser Ensembl (http://ensembl.org) has integrated Wasabi to provide interactive visualization of the Genetree datasets. To see Wasabi in action, click on any node in a genetree (e.g. BRCA2 genetree) and click “Wasabi viewer”. The selected subtree…
Wasabi integrated to ConSurf server
In addition to our public server, Wasabi can be added to other web services for customized integration. The ConSurf server is using Wasabi to provide interactive visualization for the result alignments and phylogenetic trees. Here’s an example results page with…
Wasabi update 1.2016
Have a great year in 2016! Here’s an update for you: New stuff Import genetrees and alignment blocks also from Ensembl Genomes Extended tree metadata import (EC/AC codes etc.) Licensing changed from GPL to AGPL New attribute (“check”) added to…
Wasabi article published!
Great news! The first article describing Wasabi and few of its use cases has been published in Molecular Biology and Evolution. You can now read the article online and cite it in publications: Veidenberg, A., Medlar, A. and Löytynoja,…