Wasabi can be learned by exploration and hovering the mouse over the interface elements displays useful information.
To start, open Wasabi and:
1. Import data
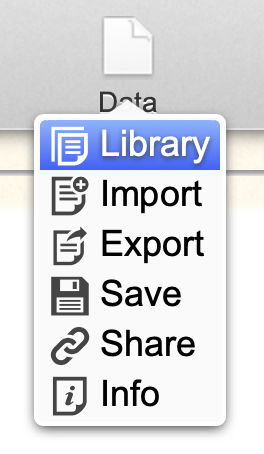
The Data menu provides several options for importing datasets:
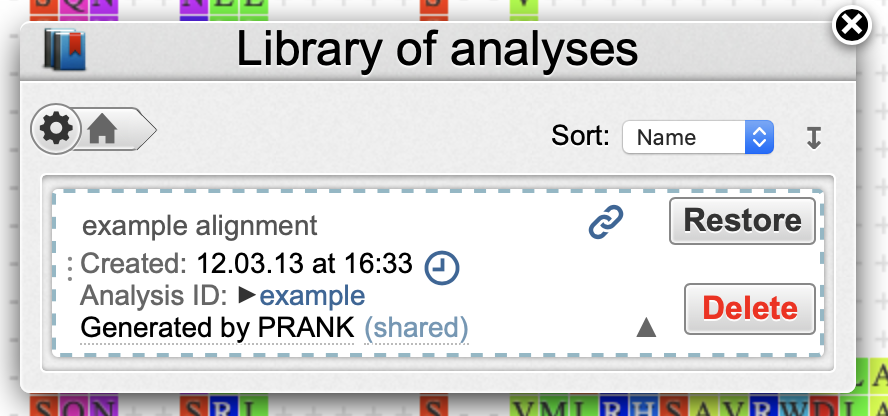
- Library gives you access to your personal analyses repository in the Wasabi server. Click Open in the Library window to load up the example dataset.
- Tip: to erase a dataset from the library, click More info → Delete.
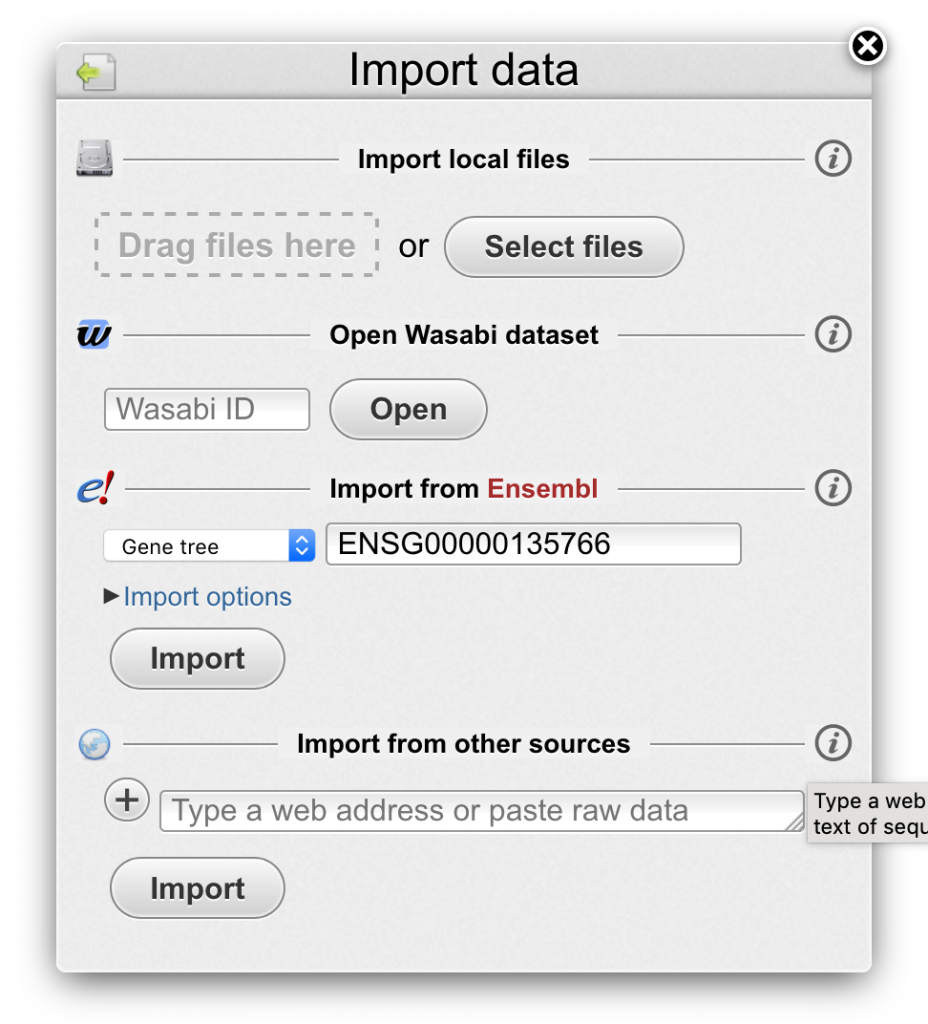
- The Import window gives access to external data sources:
- Drag files directly from a folder to the file drag area or open a file browser with Select files button.
- Wasabi expects 1-2 files containing multiple DNA/protein sequences and/or a phylogenetic tree.
- To import a shared Wasabi dataset, type its ID and click Open.
- If you have a particular gene in mind, type its name, Ensembl Gene ID or GeneTree ID in to the designated field and click Import.
- The universal input field at the bottom accepts both copy-pasted raw text or web URL to a data file in a variety of data formats.
- You can add multiple data sources and expand the input field.
- Drag files directly from a folder to the file drag area or open a file browser with Select files button.
2. Browse and edit
- Use the arrow keys, mouse scroll-wheel or drag the ruler to move around the sequence alignment.
- Drag the button above the phylogenetic tree or the divider between the tree and the sequence area to rearrange work space.
- Click on the lower-left edge of the top toolbar to hide it.
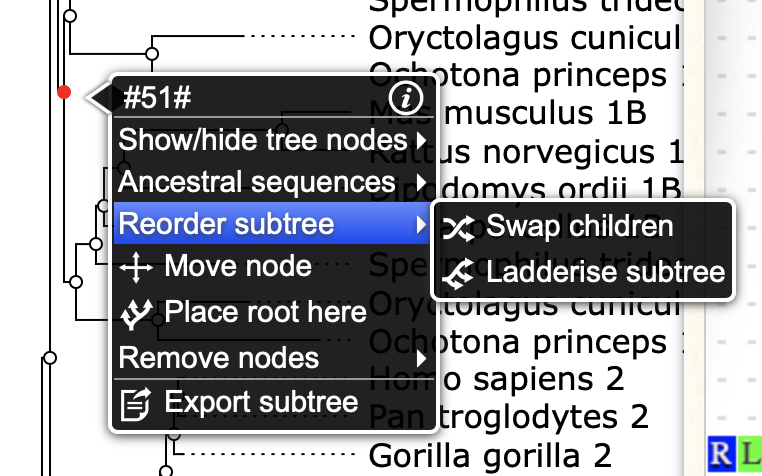
- Clicking on any tree node or leaf opens up a pop-up menu for different options for that subtree.
- You can drag/drop tree nodes to rearrange or prune branches.
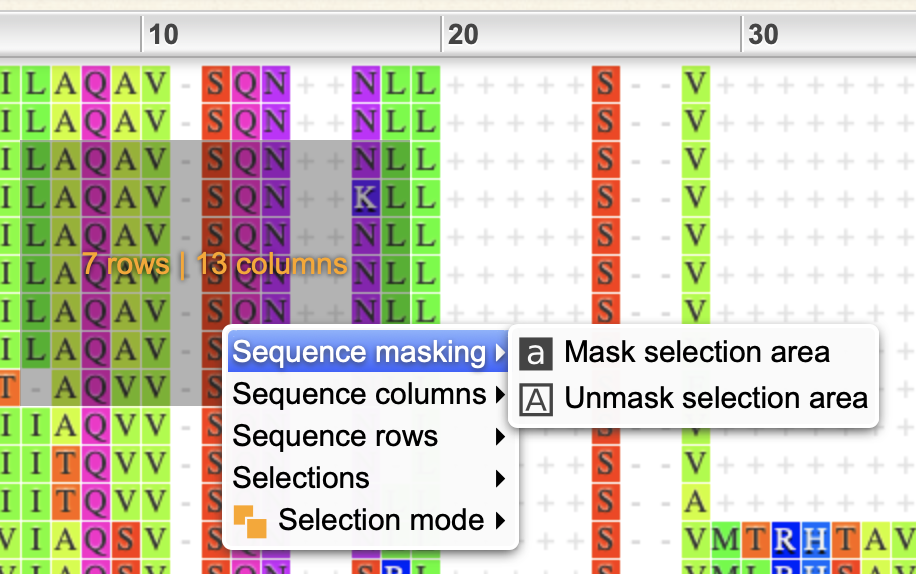
- In the sequence area, you can click for an info dialog, drag to make selections or right-click inside/outside of a selection for an appropriate pop-up menu.
- Most of edits can be undone/redone using the toolbar buttons.
- Click the button between undo/redo to see edits history and to pick a custom edit point to revert.
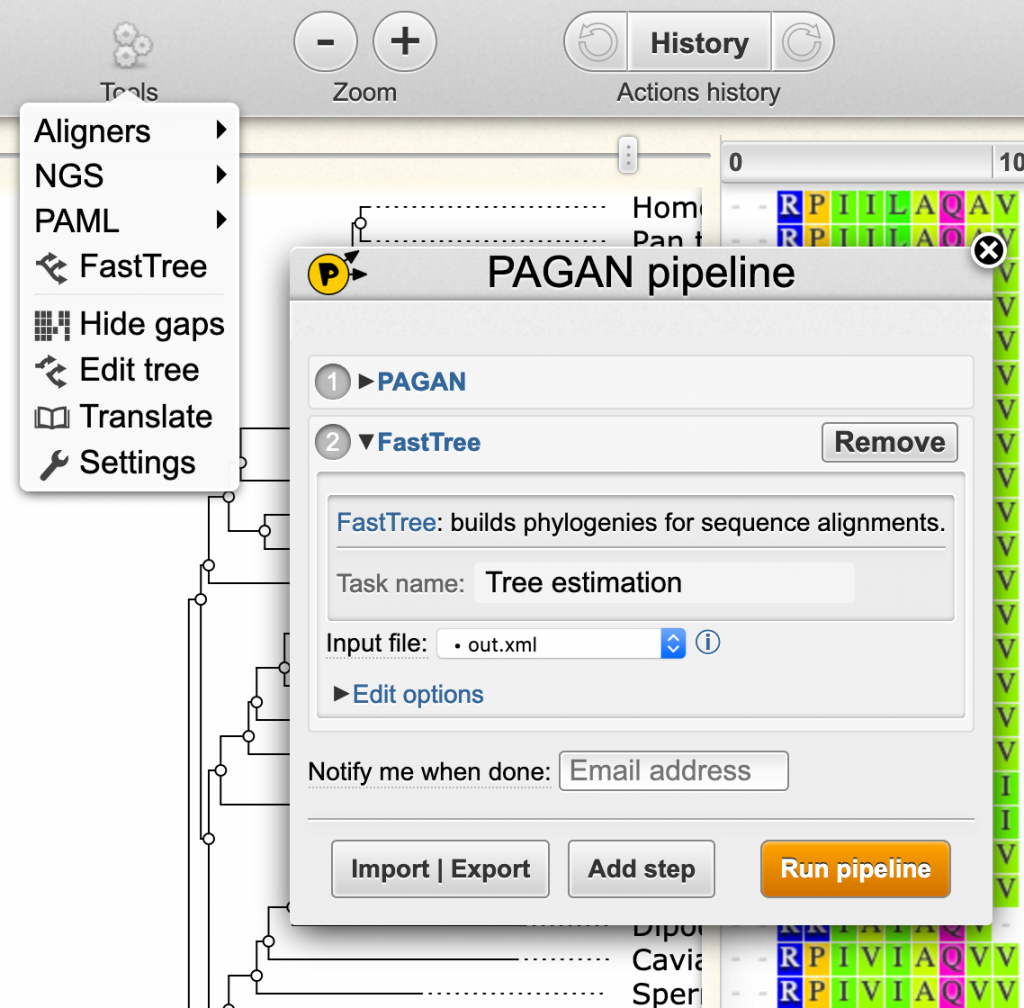
- The Tools menu provides editing tools and external analysis programs to apply for the imported dataset.
- External command-line programs are integrated to the Wasabi interface with the Pline plugins system. After selecting the analysis tool, fill the input options and click the orange launch button. The currently open dataset will be sent to the Wasabi server for analysis and a notification button will appear on the toolbar. Click the button to see the progress and to import the results.
- Visit the Pline webpage to see more plugins, pipelines and instructions.
- Interactive editing tools like sequence translation and bulk editing and annotation run inside the Wasabi interface. Edits on a specific tree node or a sequence area can be done via pop-up menus.
- External command-line programs are integrated to the Wasabi interface with the Pline plugins system. After selecting the analysis tool, fill the input options and click the orange launch button. The currently open dataset will be sent to the Wasabi server for analysis and a notification button will appear on the toolbar. Click the button to see the progress and to import the results.
3. Save data
- Data menu → Save gives you an option to store a snapshot of the current dataset to your analysis library in the Wasabi server.
- Click on the name field in the Save window, Library window or on top of the menu toolbar to edit and update it in the server.
- Data menu → Export allows you to save the dataset in a chosen file format to a local disk.